Environmental Influence on Gene Networks – Overview
Environmental Influence on Gene Networks
Overview
Introduction:
(Quick link to: Unit Plan)
In this module, students complete the steps scientists take when using systems biology to investigate how organisms induce phenotypic changes in response to the environment. Student teams apply their background knowledge of genetics and networks to experiment with a model organism. They then exchange and interpret information in order to build a possible network, and in the laboratory, test this network by altering environmental conditions. This leads to further experimentation to verify and draw conclusions about network interactions using experimental data and a computer simulation. Students act as scientists while planning, implementing, and evaluating an investigation in the context of a real regulatory network.
Pacing Guide:
 Lesson 1 – Scientists Prepare and Plan
Lesson 1 – Scientists Prepare and Plan
Gaining background knowledge, forming questions and planning experiments.
1, 50-minutes class period
Lesson 2 – Growth and Phenotypic Response of Halobacterium in Different Environmental Conditions
Benchwork, experimentation in the lab
3 class periods, including AT LEAST 72 hours for incubation
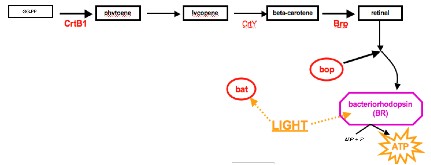
The exploration of the genetic control of networks begins as students go into the laboratory to test how gene expression in Halobacterium changes in response to environmental conditions. Surprisingly, students observe that the cells, typically pink in color, experience a phenotypic change when grown in the presence of a full spectrum light bulb and when grown in the absence of all light. The cells grown in dark conditions turn nearly white, whereas cells grown in the presence of light turn a deep purple color.

Students also notice that the amount of cell growth increases when a full-spectrum light bulb is used. After learning that bacteriorhodopsin (bR), a protein found in the Halobacterium membrane, is purple, students begin to suspect that the genes for bR might be turned on in the presence of light and turned off in the absence of light. After gathering and graphing all of the class data using Excel, they induce that as the amount of light increases, so does the amount of bR. However, they find they do not have enough information to verify this, which sends them in search of more evidence to answer the question, “How might Halobacterium cells change the amount of bR in response to light?”
Lesson 3 – Data Analysis to Propose Network Function
Collaboration with other experts to thoroughly analyze multiple types of available, pertinent data.
2 class periods
Networks of genes and proteins allow cells to change gene expression in response to their environment. Just as scientists use this premise to infer or build models of cellular networks by analyzing and integrating multiple types of data, the students likewise use actual data to piece together a network in order to answer their question discovered through experimentation. Each student analyzes one of three types of data using given background information, actual homology, metabolic or microarray data, and a worksheet to guide their analysis. The student then joins a pre-assigned group to teach each other about their data and information. Together the group uses all of their new understandings to pencil in a possible bacteriorhodopsin network. Because this would not be possible with only one type of data, this displays the significance of compiling multiple data sets from various experimental techniques when suggesting network relationships. The data also gives the students the opportunity to look past complex names to see molecules as items that simply interact. They represent these molecules as symbols, or nodes, in a network and begin to visualize how a set of proteins and genes function as a unit to give rise to a set of properties within a cell. Specifically, they learn about growth and energy production in response to changing the environmental conditions of light and oxygen; in general, they learn about systems concepts along with the importance of both gene regulation and surviving in a variety of environments.
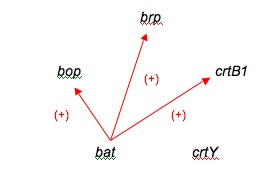
One of the beginning networks after looking at only microarray data.
Final, class compiled bR network.
Lesson 4 – Analysis of Laboratory Results to Verify Network Interactions
Scientists combine previous data with their new data in order to evaluate their proposed network function.
3 periods
Once the students begin to understand the make up of their bR network as well as the negative and positive relationships within the network, they realize the network in this stage is actually a hypothesis. They consider how to test this model, which brings them back to the lab to revisit their initial question of, “How might Halobacterium cells change the amount of bR in response to light?” Students test the validity of their models/hypotheses by comparing predicted phenotypes of genetically manipulated strains with observed phenotypes. Their predictions come from analyzing their proposed network to guess what phenotypes would be present when different routes within their network are active. For instance, if the model is correct, then when the bat gene is knocked out, the cells should look a certain way. The students actually spin down cells of varying strains, with known genotypes, to view the cultures’ specific phenotypes. To bring this back to their original question, they gather the cell pellets they created when growing wild type Halobacterium in different conditions (including light and dark conditions) and compare them to the different mutant strains.
By matching the phenotypes of the experimental and known cell pellets using a color chart, they can infer what network interactions are currently at play. This information is used to answer their question and verifies how a certain environmental condition affects gene expression and the cellular network. Students further their understanding by manipulating a computer simulation built by scientists in the Baliga Lab to display the actual bR network.

Many important systems concepts come from these lessons. Please see our Unit Plan for more information. A strength of this module, is its ability to have students actually view how a response to a stimulus occurs by changing the relative amount or function of individual nodes in a cellular network. They can further see how that stimulus reverberates throughout a network – it is very apparent when studying their bR network that one node can cause a change in many other nodes due to the interconnected nature of a network. It is also very apparent that some nodes have bigger system-wide effects than others. By bringing in mutant strains and discussing the relevance of experiments such as these to real-world situations, students can also begin to understand how the relative importance of nodes allows for greater system understanding. This leads to deep comprehension of how to perturb and use the system, which has numerous applications in science and healthcare.
All of our curricula, worksheets, formative and summative assessments are built into web-based lesson plans complete with needed presentations, background information and laboratory materials. Please see subsequent pages for more information.
For background information emphasizing the need for basic research, please see this short article on the importance of studying bacteriorhodpsin. This was taken from the August 2011 HHMI Bulletin, vol. 24, No. 3. “In Support of Undirected Research” by Karl Deisseroth.
Module FLOW CHART
One-page description of Environmental Influence on Gene Networks module


